
Nonetheless, we are still in the very early phases of designing artificial proteins for specific functional purposes. Consequently, some groups have steered away from utilizing natural protein scaffolds as a starting point for engineering, and instead create completely artificial, human-made proteins ( Lombardi et al., 2000 Faiella et al., 2009 Anderson et al., 2014 Solomon et al., 2014). Thus, epistatic relationships between residues pose a large challenge to protein engineers who seek to achieve more than a modest improvement in protein activity. For example, the evolutionary trajectory of influenza nucleoprotein revealed that stabilizing substitutions were essential in compensating for otherwise intolerable destabilizing changes that were beneficial to the protein in an adaptive sense ( Gong et al., 2013). Indeed, historical contingency and entrenchment due to epistasis play a critical role in protein evolution ( Weinreich et al., 2006 Bridgham et al., 2009 Bloom et al., 2010 Gong et al., 2013 Harms and Thornton, 2014 Shah et al., 2015). These various historical constraints and complexities can confound engineering approaches. Moreover, natural proteins may also have been subjected to multiple, simultaneous and diverse selection pressures leading to complexity from Darwin's principle of multiple utility ( Dutton and Moser, 2011). Amino acids within natural proteins may display Müllerian interdependency and consequently may have accumulated irreversible complexity ( Dutton and Moser, 2011).
Understanding the basis for these constraints has proven challenging, as evolution has sculpted epistatic relationships between residues and structural domains in proteins that are necessary for thermodynamic stability, dynamics, allostery, or complex assembly ( Liberles et al., 2012 Gong and Bloom, 2014 Perica et al., 2014 Sikosek and Chan, 2014). From engineering DNA polymerases to better recognize unnatural nucleotides ( Laos et al., 2014), to introducing catalytic activity into non-enzymatic scaffolds ( Korendovych et al., 2011), protein engineering has enhanced our repertoire of knowledge far beyond solely a fundamental understanding of protein structure and function.ĭespite such progress, there are major challenges to protein engineering, mostly rooted in the abundance of constraints embedded in natural protein sequences and structures ( Dutton and Moser, 2011). A wide spectrum of proteins have been successfully engineered and evolved to adopt new or improved function. An exciting assortment of approaches has been developed to optimize starting protein scaffolds, ranging from the development of site-directed mutagenesis in the early 1980s ( Dalbadie-Mcfarland et al., 1982 Sigal et al., 1982 Winter et al., 1982), to more recent computational design efforts ( Jiang et al., 2008 Siegel et al., 2010). Protein-engineering endeavors have met both challenge and opportunity in designing proteins with desirable or novel functions. The advancement of protein engineering has enriched our understanding of protein structure, function, and folding. They also suggest that enhanced chaperones and disaggregases could have important applications in treating human disease as well as in the purification of valuable proteins in the pharmaceutical sector. Collectively, these advances have revealed key mechanistic and functional insights into chaperone and disaggregase biology. Likewise, small changes in the primary sequence of Hsp104 yield potentiated protein disaggregases that reverse the aggregation and buffer toxicity of various neurodegenerative disease proteins, including α-synuclein, TDP-43, and FUS. Remarkably, enhanced global activity or altered substrate specificity of various molecular chaperones, including GroEL, Hsp70, ClpX, and Spy, can be achieved by minor changes in primary sequence and often a single missense mutation. Here, we review efforts to enhance the activity of individual molecular chaperones or protein disaggregases via engineering and directed evolution. A promising strategy is to engineer proteostasis networks to counter challenges presented by specific diseases or specific proteins. Nevertheless, proteostasis networks have limits, which when exceeded can have fatal consequences as in various neurodegenerative disorders, including Parkinson's disease and amyotrophic lateral sclerosis.

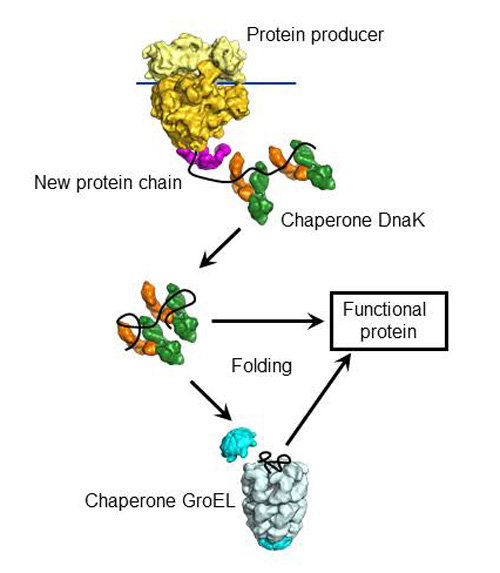
Critical components of this network include molecular chaperones and protein disaggregases, which function to prevent and reverse deleterious protein misfolding.
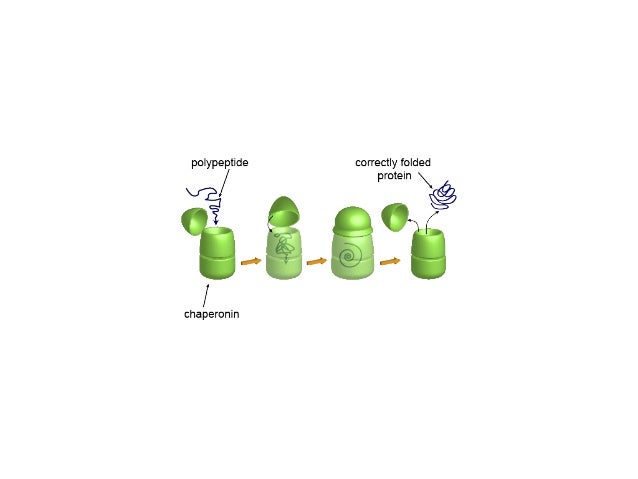
2Biochemistry and Molecular Biophysics Graduate Group, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, USAĬells have evolved a sophisticated proteostasis network to ensure that proteins acquire and retain their native structure and function.1Department of Biochemistry and Biophysics, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, USA.


 0 kommentar(er)
0 kommentar(er)
